Adverse Drug Reactions (ADRs) are the fourth leading cause of death in the US. One such cause of ADRs is brought about through Drug-Drug Interactions (DDIs). The positive side of this is that such reactions can be prevented.
Information related to DDIs is dispersed across different biomedical articles and is growing at an accelerant rate. Currently there are a number of free repositories available online, such as DrugBank and Drugs.com that store information about known DDIs. Nonetheless, these repositories feature a limited amount of such DDIs and they are only updated every two years. For this reason, we propose medicX, presented in Figure 1, a system that is able to detect DDIs in biomedical texts for healthcare professionals, by leveraging on different machine learning techniques.
The main components within our system are the Drug Named Entity Recognition (DNER) component that identifies drugs within the text, and the DDI Identification component that detects interactions between the identified drugs. Different approaches were investigated in line with existing research. The DNER component is evaluated using the BioCreative CHEMDNER [1] and the DDIExtraction 2013 [2] challenge corpora. On the other hand, the DDI Identification component is evaluated using the DDIExtraction 2013 [2] challenge corpus. The DNER component is implemented using an approach based on bi-directional Long Short-Term Memory (LSTM) networks with Conditional Random Fields (CRF). The LSTMs are used to learn word and character based representations from the biomedical texts, whilst the CRFs are used to decode these representations and identify drugs among them.
This method achieves a macro-averaged F1-score of 84.89% when it is trained and evaluated on the DDI-2013 corpus, which is 1.43% higher than the system that placed first in the DDIExtraction 2013 challenge [3]. Furthermore, our approach is efficient because it is able to identify drugs in sentences instantly and it does not require any additional lexical resources.
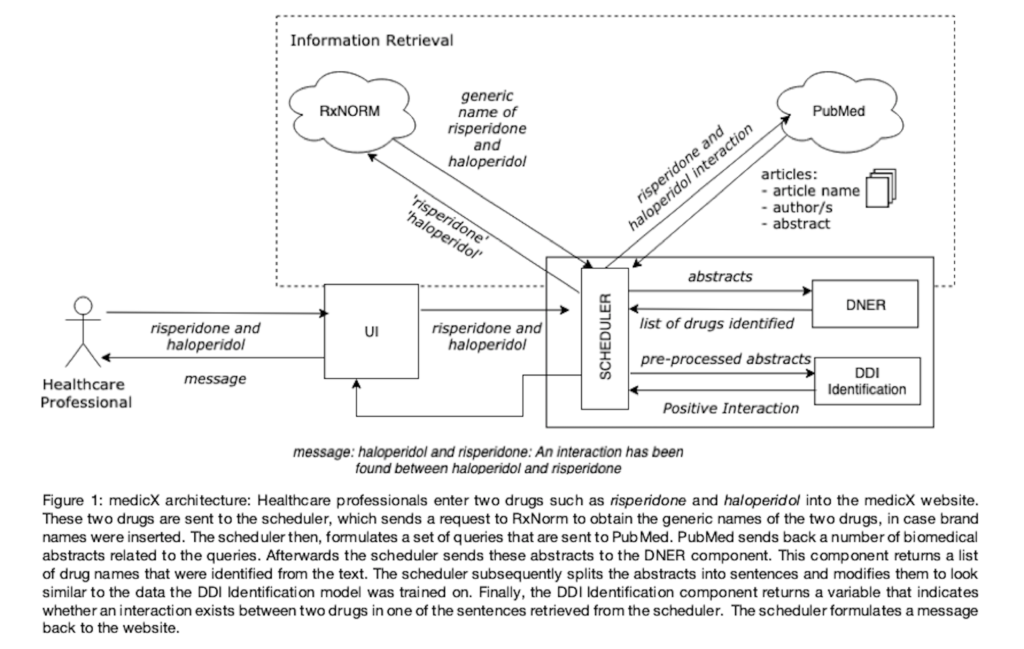
On the other hand, the DDI Identification component is implemented using a two-stage rich feature-based linear-kernel Support Vector Machine (SVM) classifier. We demonstrate that calculating the average word embedding of a sentence and detecting trigger words in sentences are rich features for our SVM classifier. Our DDI Identification system achieves an F1-score of 66.18%, as compared to the SVM state-of-the-art DDI system that reported an F1-score of 71.79% [4]. Moreover, when our system was evaluated on a subset of this corpus that consisted solely of long and complex MedLine sentences, our system came second, following the state-of-the-art DDI system developed by Zheng et al. [5] that uses neural networks to locate DDIs. Our system shows very encouraging results.
References
[1] Krallinger, M., Rabal, O., Leitner, F., Vazquez, M., Salgado, D., Lu, Z., Leaman, R., Lu, Y., Ji, D., Lowe, D.M. and Sayle, R.A., 2015. The CHEMDNER corpus of chemicals and drugs and its annotation principles. Journal of cheminformatics, 7(1), p.S2.
[2] Herrero-Zazo, M., Segura-Bedmar, I., Martínez, P. and Declerck, T., 2013. The DDI corpus: An annotated corpus with pharmacological substances and drug–drug interactions. Journal of biomedical informatics, 46(5), pp.914-920.
[3] Rocktäschel, T., Huber, T., Weidlich, M. and Leser, U., 2013. WBI-NER: The impact of domain-specific features on the performance of identifying and classifying mentions of drugs. In Second Joint Conference on Lexical and Computational Semantics (* SEM), Volume 2: Proceedings of the Seventh International Workshop on Semantic Evaluation (SemEval 2013) (Vol. 2, pp. 356-363).
[4] Raihani, A. and Laachfoubi, N., 2017. A rich feature-based kernel approach for drug-drug interaction extraction. International journal of advanced computer science and applications, 8(4), pp.324-3360.
[5] Zheng, W., Lin, H., Luo, L., Zhao, Z., Li, Z., Zhang, Y., Yang, Z. and Wang, J., 2017. An attention-based effective neural model for drug-drug interactions extraction. BMC bioinformatics, 18(1), p.445.
1 https://bit.ly/2vaWF6e, accessed on 05/12/2018
2 https://www.drugbank.ca accessed on 15/07/2018
3 https://www.drugs.com/ accessed on 15/07/2018
4 https://rxnav.nlm.nih.gov/RxNormAPIs.html, accessed on 30/11/2018 5 https://www.ncbi.nlm.nih.gov/pubmed/ accessed on 13/10/2018
Student: Lizzy Elaine Farrugia
Supervisor: Dr Charlie Abela
Course: B.Sc. IT (Hons.) Artificial Intelligence
