Biological curation is the cornerstone of modern Computational Biology. Specialised Biocurators can be likened to the museum catalogers in the world of Computational Biology. They turn unidentifiable objects from unwieldy mediums into a powerful model from which researchers can benefit from[1].
Our hypothesis premises that through property graphs, it is possible to model properties of ontologies such that they can be used by text mining algorithms to find appropriate ways to formally describe complex interactions in biology. Moreover, we hypothesise that it is possible to use domain-specific ontologies to describe such relations in biological literature.
In order to achieve our aims we obtained a number of ontologies from OBO Foundry, which is a repository of biological ontologies that are logically well-formed and scientifically accurate[2]. A subset of interactions in our Knowledge Graph were validated against the Chemical Disease Relationship(CDR) dataset[3]. The CDR dataset is a curated set of PubMed abstracts describing chemical to disease interactions.
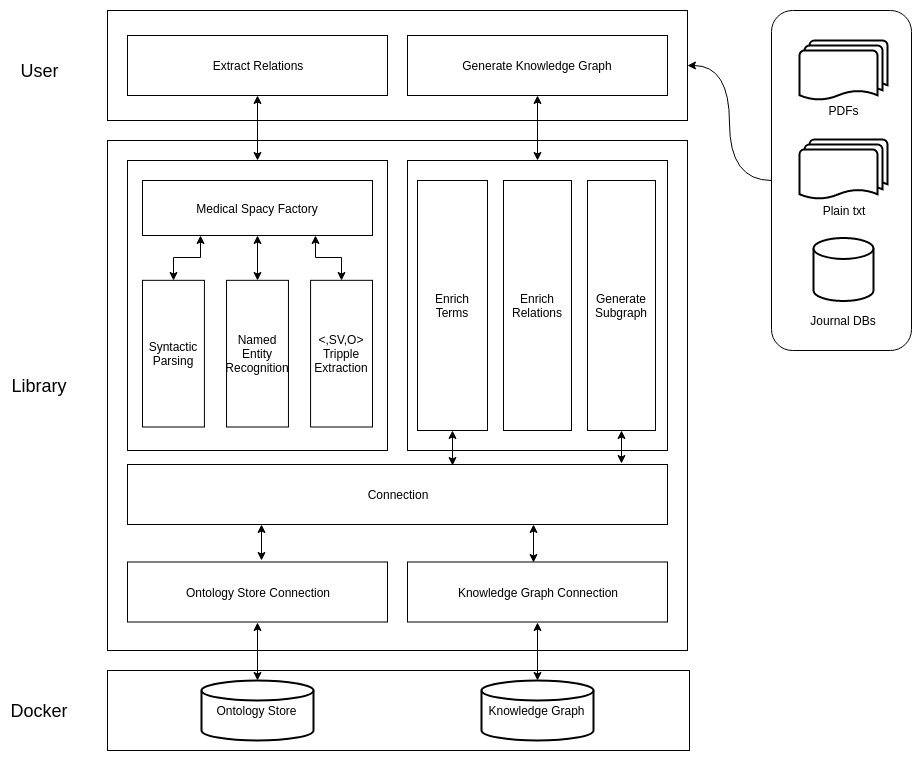
We created a Python library that encapsulated an NLP pipeline to automatically extract Subject-Verb-Object tuples as shown in Fig 1. The subjects and objects are further enriched by getting the best term representing them from the Ontology
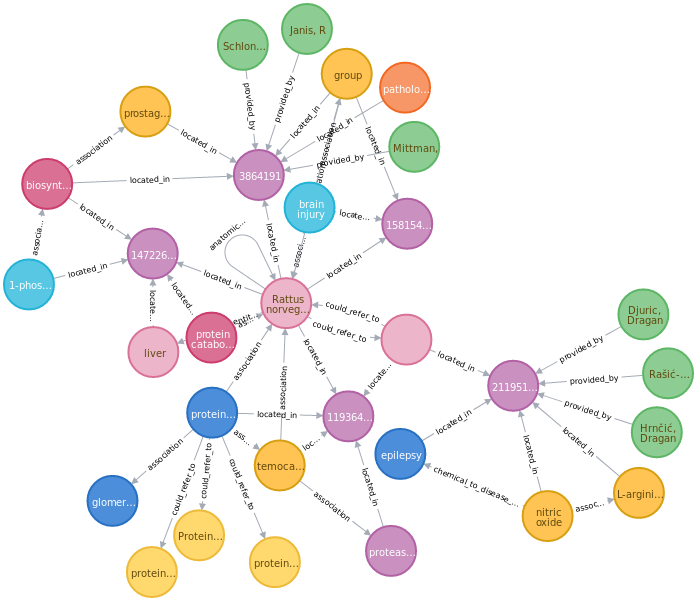
Store. Relations whose subject and object are represented in the Ontology Store are represented in our Knowledge Graph as shown in Fig 2. The Knowledge Graph uses a standard defined by Biolink as a ‘schema’. The graphs were stored and represented using two Neo4j graph databases orchestrated through Docker and Docker Compose.
When compared to the CDR dataset, we managed to achieve an F-Score of 0.25 which is within the baseline margin of error. There were specific tools that managed to obtain a better score. However, this result is encouraging and we believe that with further tweaking we can improve this score significantly.
Whilst our system obtained scores close to the task benchmark it requires further enhancements to obtain scores similar to other teams in the competition. Our scores would improve if we:
- Replace the <S,V,O> extraction utility with Semantic Role Labelling;
- Represent ontologies in an RDF Triple Store instead of Neo4j in order to take advantage of Description Logic and Ontology Axioms;
- Investigate integrating Elasticsearch for resolving classes from ontologies.
References
[1] P. E. Bourne and J. McEntyre, “Biocurators: contributors to the world of science.,” PLoS computational biology, vol. 2, no. 10, e142, 2006, issn: 1553-358. doi: 10.1371/journal.pcbi.0020142. [Online]. Available: http://www.ncbi.nlm.nih.gov/pubmed/17411327 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC1626157
[2] B. Smith, M. Ashburner, C. Rosse, J. Bard, W. Bug, W. Ceusters, L. J.Goldberg, K. Eilbeck, A. Ireland, C. J. Mungall, t. O. OBI Consortium, N. Leontis, P. Rocca-Serra, A. Ruttenberg, S.-A. Sansone, R. H. Scheuermann, N. Shah, P. L. Whetzel, and S. Lewis, “The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration.,” Nature biotechnology, vol. 25, no. 11, pp. 1251–5, 2007, issn: 1087-0156. doi: 10 . 1038 / nbt1346. [Online]. Available: http://www.ncbi.nlm .nih.gov/pubmed/17989687 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC2814061.
[3] C.-H. Wei, Y. Peng, R. Leaman, A. P. Davis, C. J. Mattingly, J. Li, T. C.Wiegers, and Z. Lu, “Overview of the BioCreative V Chemical Disease Relation (CDR) Task,” Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, pp. 154–166, 2015.
Student: Matthew Drago
Supervisor: Mr Joseph Bonello
Co-Supervisor: Prof. Ernest Cachia
Course: B.Sc. IT (Hons.) Software Development
