This bioinformatics-oriented project has tackled the task of classifying amyloidogenic hexapeptides. Amyloids are proteins that may form fibrils instead of functioning as intended. These proteins may build up in organs such as the heart, kidneys, liver, and brain, and
are responsible for a group of diseases referred to as amyloidosis, which includes Alzheimer’s disease, Parkinson’s disease and Type II diabetes.
In essence, a protein consists of one or more chains of amino acids called polypeptides. In the field of bioinformatics, polypeptides could be represented as sequences of amino acids, where each amino acid is represented by its unique symbol. In fact, the amino acid alphabet contains 20 symbols, each representing a different amino acid. A hexapeptide is a chain of six amino acids, and it has been shown that short chains of six amino acids may be responsible for amyloidogenicity. Hence, these were sufficient for the classification problem at hand. This is particularly significant, as the majority of datasets of labelled peptides publicly available are hexapeptide datasets (datasets of labelled chains of six amino acids).
It is currently too expensive and time consuming to experimentally assert all possible sequences of hexapeptides as amyloid or non-amyloid. Therefore, it would be useful for researchers to be able to rely on computational models that could classify hexapeptides in a short amount of time, aiding them in their research on these diseases. Indeed, this project focused on investigating the application of grammatical inference techniques to this problem.
Grammatical inference is a field of study where a formal language would be inferred from a set of example strings that belong to a language, and a set of counterexamples that do not belong to the
language. In simpler terms, grammatical inference techniques infer a set of ‘rules’ ‒ usually in the form of a mathematically constructed abstract machine ‒ that could be used to classify a given sequence of amino acids symbols as amyloid or non-amyloid.
The task was accomplished by first researching the field of grammatical inference in order to gain a solid background of the fundamentals of the field. The next step was an assessment of other
studies that used the grammatical inference approach to solve this problem, with a view to replicating their experiments. With acquired background knowledge of the field it was possible to undertake an interpretation of the results obtained in the replicated studies
The final step in the project consisted in comparing grammatical inference techniques implemented with more traditional machine learning methods, with the aim of identifying any differences in performance among the different methods.
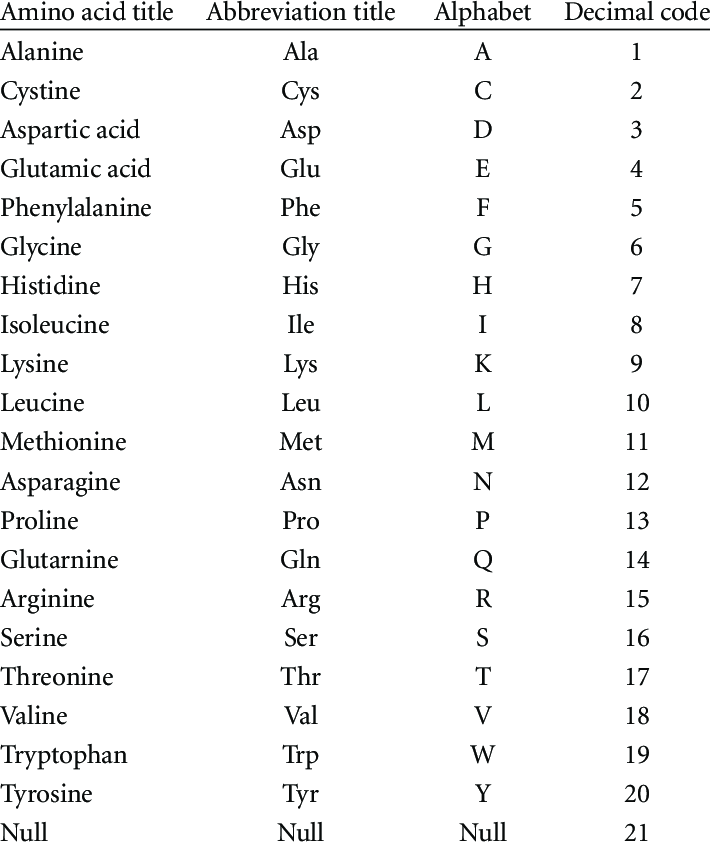
symbols [1]
References/Bibliography:
[1] Z. Zheng, Y. Chen, L. Chen, G. Guo, Y. Fan, και X. Kong, ‘Signal-BNF: A Bayesian Network Fusing Approach to Predict Signal Peptides’, Journal of biomedicine & biotechnology, τ. 2012, σ. 492174, 10 2012.
Student: Malcolm Agius
Course: B.Sc. IT (Hons.) Artificial Intelligence
Supervisor: Dr Kristian Guillaumier
