Biologists frequently need to combine information from different databases by manually following hyperlinks between different databases and using the distinct interfaces provided by the different
databases. This is cumbersome and time consuming. Moreover, biologists require information on different aspects of proteins such as the protein structure, sequence, function, and its interactions with other biomolecules.
To alleviate this, past research in data integration involved attempts to provide a single access point for all required biological data. One approach is “Data Warehousing”, where data from different databases is combined within a centralised repository. However, the current state
of the art in biological data warehousing requires bespoke software development and maintenance for each database. This is infeasible since data is spread over many databases which are constantly changing.
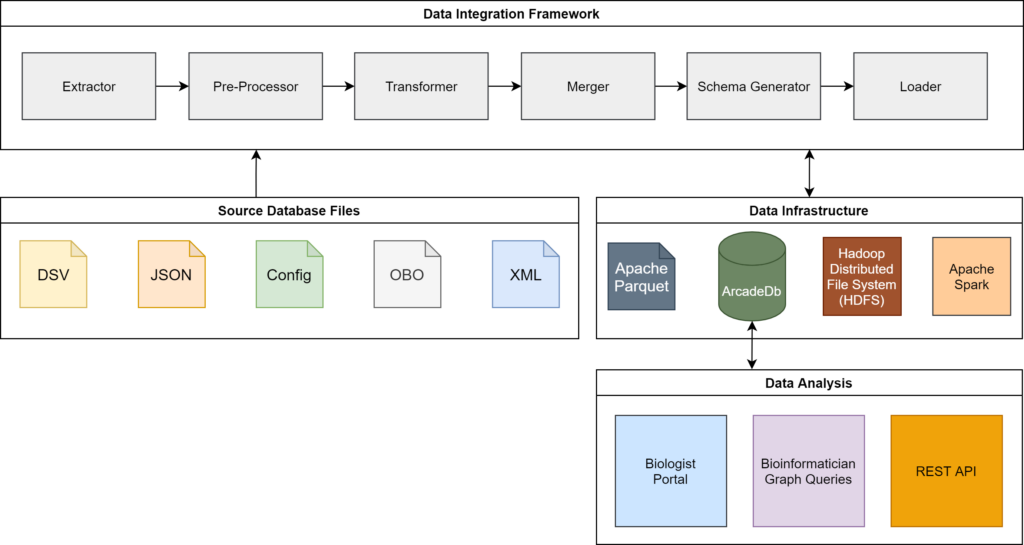
This research aims to eliminate this process through a semi- automated data integration system. Given user-defined configurations, the prototype developed can automatically retrieve
information from the original databases and load it in a graph database. Using Apache Spark and the Hadoop Distributed File System (HDFS) enables horizontal scalability within the “Pre-
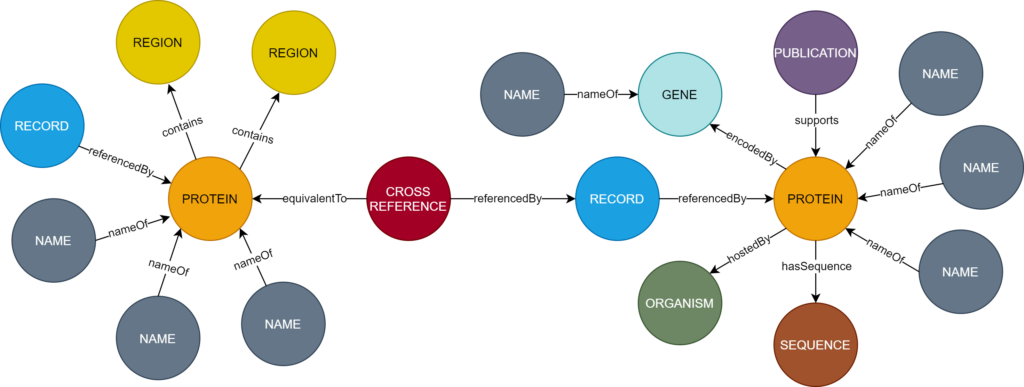
Processor”, “Transformer” and “Merger” components. The system offers a user interface which presents all identified information for a protein in a unified manner. The graph database also allows technical users to code for more complex questions such as “Which proteins are known to interact with proteins involved in Alzheimer’s disease?”
The prototype was applied to a restricted set of protein databases and file formats due to time and space constraints. However, the results are promising and with further development, the tool can improve the time required to find protein information. The next steps include extending the “Transformer” module to handle further cases, improving the performance for retrieving structural domains, and applying the tool to more databases and file formats.
Student: Jurgen Aquilina
Course: B.Sc. IT (Hons.) Software Development
Supervisor: Mr Joseph Bonello
