A brain haemorrhage is a rupture of the blood vessels within the brain, and is very often life-threatening [1]. Brain haemorrhaging is the third-leading cause of mortality across all age groups and is mainly caused by haemorrhagic stroke or trauma [2]. There are various types of blood haemorrhage, including: intracerebral, intraventricular, intraparenchymal, subarachnoid, subdural, and epidural haemorrhages.Diagnosis tools used by medical experts to identify the type of pathology include: computed tomography (CT) or magnetic resonance imaging (MRI) scans, lumbar puncture, or cerebral angiography [3]. This study focuses on the use of CT scans as a method for diagnosis.
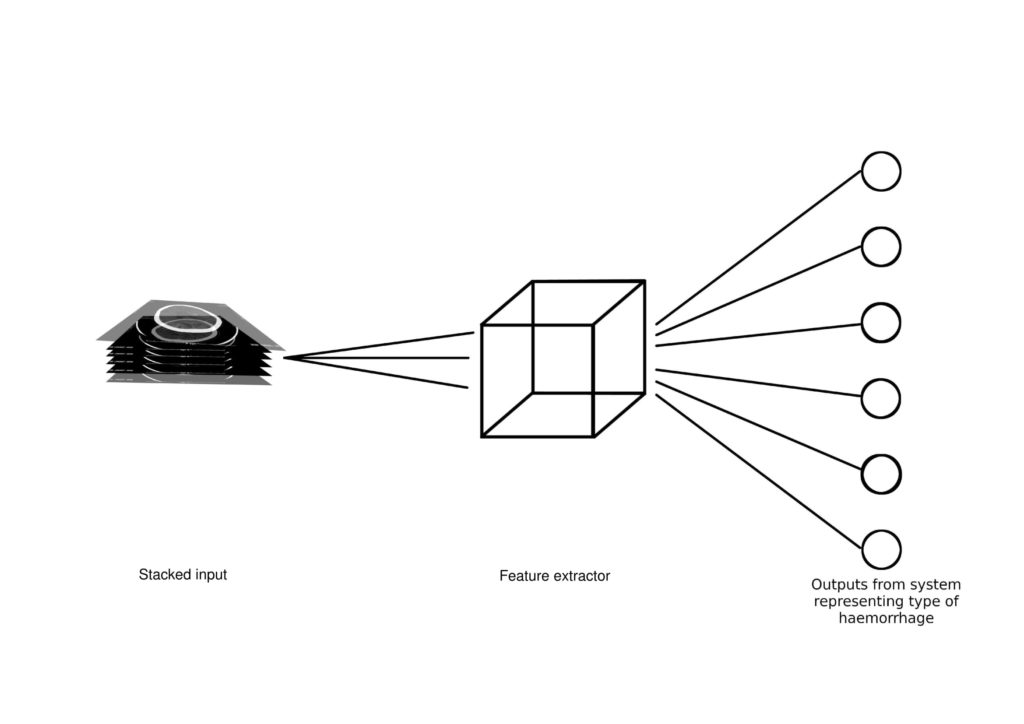
The detection and localisation of a brain haemorrhage is highly time-critical, as the longer a case goes undiagnosed, the higher the possibility of a fatality [4]. This study focuses on developing an automated software tool to aid radiographers in classifying the type of bleeding present in a series of CT scan slices, and hence localising it. This is accomplished through the design of a computer-aided diagnosis (CAD) system based on a deep learning algorithm. Specifically, this study explores the use and implements a three-dimensional convolutional neural network (3D CNN). A CNN is a network of layers that reduce an image to its most basic features, making classification easier. The convolutional layer is the layer that translates the image to usable data, scanning
small sections of the image and assigning them to different filter classes. The same holds for a 3D CNN, except that in this case the kernels move through three dimensions of data and produce 3D activation maps [5].
When building a CNN structure, it is always best to start with as small a setup as possible and gradually expand, increasing layers and units until the validation error stops improving. Architectural optimisations were also used in this study to improve computational performance. These included the use of skip connections and pointwise filters.
To train and test the developed solution, this study makes use of a dataset of 143 cases, where each case holds multiple slices of CT scan images. In preparation for the next stage, each image was reduced to a 128×128 pixel image and stacked, in order to obtain 143 stacked 3D images with a shape of 100×128×128×1, where 1 represents the greyscale channel and 100 is the stack height. In cases that did not contain precisely 100 images, the available images were truncated or padded accordingly. During this process an image augmentation was also undertaken so as to obtain another 65 images for each training case. The dataset was split into: 80% training and 20% for testing. The divide was carried per classification, rather than as a whole, to ensure a proportionate distribution. The derived results obtained were deemed satisfactory for the purposes of the project.
References/Bibliography:
[1] “Cerebral Hemorrhage,” accessed 27th August 2020. [Online]. Available: https://www.merriam-webster.com/medical/cerebral%20 hemorrhage
[2] N. Spiteri, “Classification of brain haemorrhage in head ct scans using deep learning,” Final Year Project, University of Malta, 2019.
[3] A. Felman, “What to know about brain hemorrhage,” 2019, accessed 27th August 2020. [Online]. Available: https://www. medicalnewstoday.com/articles/317080
[4] M. Grewal, M. M. Srivastava, P. Kumar, and S. Varadarajan, “RADNET: radiologist level accuracy using deep learning for HEMORRHAGE detection in CT scans,” CoRR, vol. abs/1710.04934, 2017. [Online]. Available: http://arxiv.org/abs/1710.04934
[5] “Understanding a 3D CNN and Its Uses,” accessed 27th August 2020. [Online]. Available: https://missinglink.ai/guides/ convolutional-neural-networks/understanding-3d-cnn-uses/
Student: John Parnis
Course: B.Sc. IT (Hons.) Computing Science
Supervisor: Prof. Ing. Carl James Debono
Co-supervisor: Dr. Paul Bezzina and Dr. Frank Zarb
