The human body keeps blood glucose levels within safe limits naturally. However, this is not the case for persons with diabetes, raising the need for blood glucose level control. This, in turn, entails regularly measuring blood glucose levels and, depending on the identified level, the appropriate treatment would then be administered.
Diabetes is a global health problem, and the International Diabetes Federation (IDF) reports that around 463 million persons are currently living with diabetes, amounting to roughly 1 in 11 adults worldwide. The IDF projects that by 2030 more than 578 million individuals will be living with diabetes [1]. Diabetic patients are at higher risk of critical glycaemic events, and such events could be mitigated through timely intervention. However, these preventive actions tend to take time to act.
The use of machine learning (ML) techniques for predicting blood glucose levels is an area of interest to researchers attempting to provide the patient with predictions on future blood glucose levels using less invasive measures than are currently in use. In order to achieve this, an adequate dataset would be required.
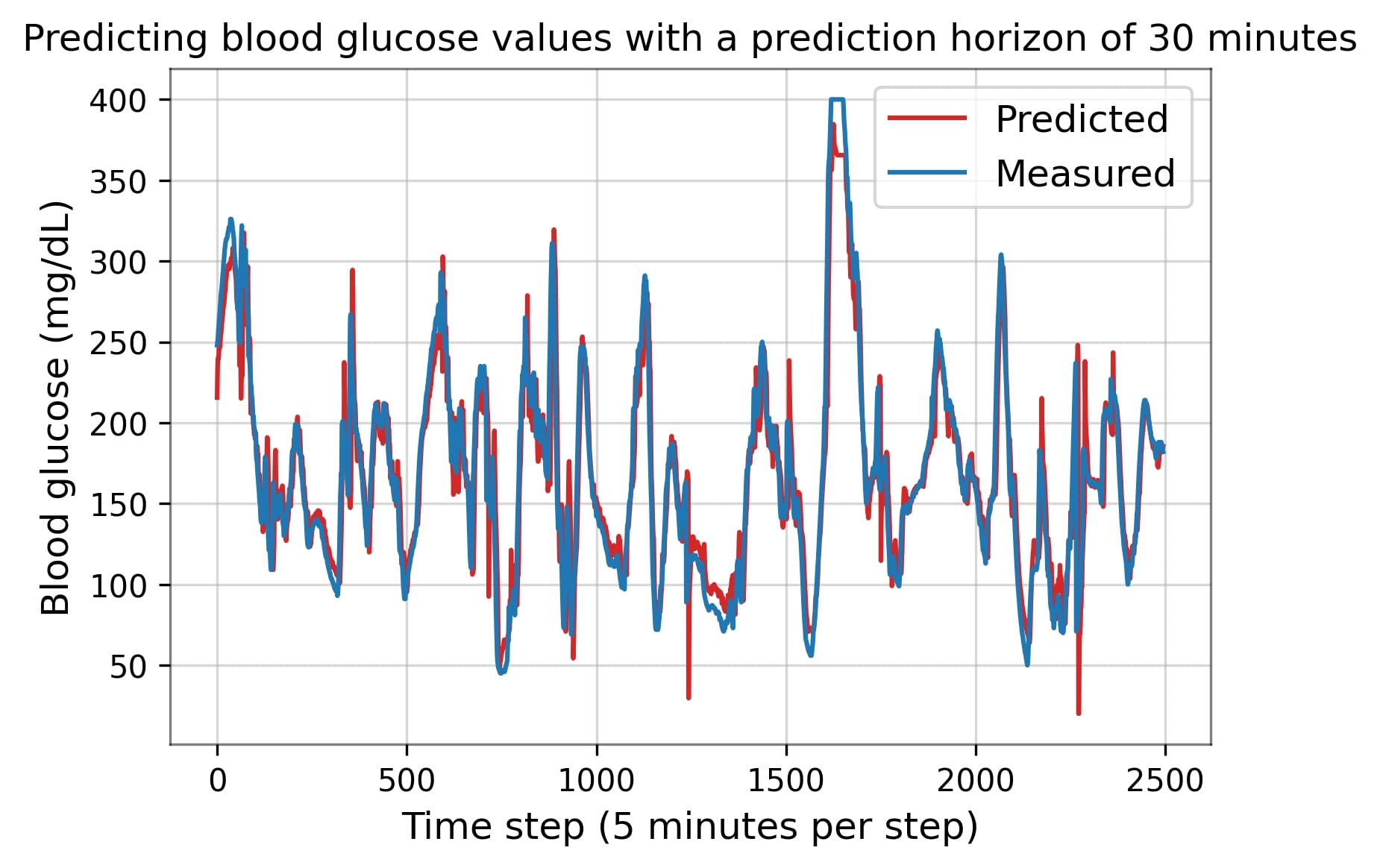
The OhioT1DM dataset is a publicly available dataset, consisting of physiological time series data of a large number of diabetes patients collected over eight weeks [2]. The dataset contains thousands of rows and multiple columns, obtained from wearable sensors and a continuous glucose monitor (CGM). In a number of related studies, multiple ML approaches were proposed for blood glucose prediction, such as the recurrent neural network and the multi-layer perceptron. This study implements two or more of these predictive algorithms using the OhioT1DM dataset, with the aim of predicting up to one hour ahead with clinically acceptable predictive accuracy.
The performance of ML algorithms is dependent on their parameter configuration or hyperparameters. The manual selection of hyperparameters can often be challenging due to the large, if not infinite, parameter search spaces. Researchers often take an ad hoc approach in seeking an acceptable combination of hyperparameters that could give satisfactory results. Given the vast number of combinations in the hyperparameter search space of certain algorithms, it is often considered unfeasible to test every combination to determine the optimal combination. Hyperparameter optimisation may be used to algorithmically select and tune the hyperparameters of an ML model. Metaheuristic optimisers use intelligent ways of exploring the search space to identify near-optimal solutions using exploration and exploitation mechanisms. The aim of this study was to apply more than one metaheuristic optimiser to the task of finding candidate combinations of hyperparameters that could possibly offer better predictive accuracy.
Although intelligently searching the search space would reduce computational costs, the population-based optimisers used would still require substantial computing resources to complete in a timely manner. This work intended to use graphic processing units (GPUs) to accelerate the training process, due to their high computational power. However, in certain cases, using a single machine alone would not be sufficient. The workload might need to be distributed among a network of machines. Hence, the study also explored the use of distributed systems to train the various models with different hyperparameter combinations in parallel, on a cloud infrastructure.
References/Bibliography
[1] “IDF DIABETES ATLAS Ninth Edition 2019.” https://diabetesatlas.org/en/
[2] C. Marling and R. Bunescu, “The OhioT1DM dataset for blood glucose level prediction: Update 2020,” CEUR Workshop Proc., vol. 2675, pp. 71–74, 2020.
Course: B.Sc. IT (Hons.) Software Development
Supervisor: Dr Michel Camilleri
Co-supervisor: Mr Joseph Bonello
