Diabetes mellitus – commonly known as diabetes – is one of the most prevalent challenges in modern healthcare. Whilst presenting itself in a variety of types, its salient characteristic is the inability of the body to regulate blood sugar (glucose) levels, significantly hindering the individual’s health and general well-being. Although a cure is yet to be found, diabetes could be kept in check through regular exercise and a well-balanced diet, coupled with keeping track of fluctuations of blood glucose through constant monitoring. The latter is considered by many as a necessary evil, as current monitoring solutions entail highly invasive procedures alongside other drawbacks that further tarnish the patient’s quality of life [1]. Recent studies have presented interesting use-cases for machine learning (ML) algorithms to predict glucose levels within a certain time frame [2, 3].
The study was carried out within the framework of two main considerations. The first seeks to establish whether physiologic parameters, gathered from non-invasive sources, could be used to improve glucose predictive accuracy. The second consideration investigates whether the elimination of data gathered in an invasive manner would yield clinically acceptable results.
Multiple data analyses were conducted using time series data obtained from a sizeable ML dataset called OhioT1DM [4]. The first phase consisted of analysing and generating predictions with the aim of improving predictive performance. This was achieved by using feature-engineering techniques and splitting the dataset into different feature combinations. Data features were organised into the following groups: Glucose (G), Insulin pump (P), Fitness band (B) and Self-reported (S). Separate analytic steps (pipelines) were performed on each group, with the aim of producing a refined feature set, upon which ML models could be applied. Different combinations were tested on multiple linear regression (MLR) and XGBoost models. The results produced were evaluated to gauge the effectiveness of the input combination. In the second phase of experimentation, glucose-level values and their derived attributes were omitted completely from input features, and the corresponding predictive accuracy was evaluated. This was done to support, or otherwise, the ongoing research objective that non-invasive glucose monitoring could be achieved by means of multi- physiologic sensor monitoring. Root mean squared error (RMSE), mean absolute relative difference (MARD), R2 coefficient, and surveillance error grid (SEG) analysis were used as metrics to evaluate the produced results.
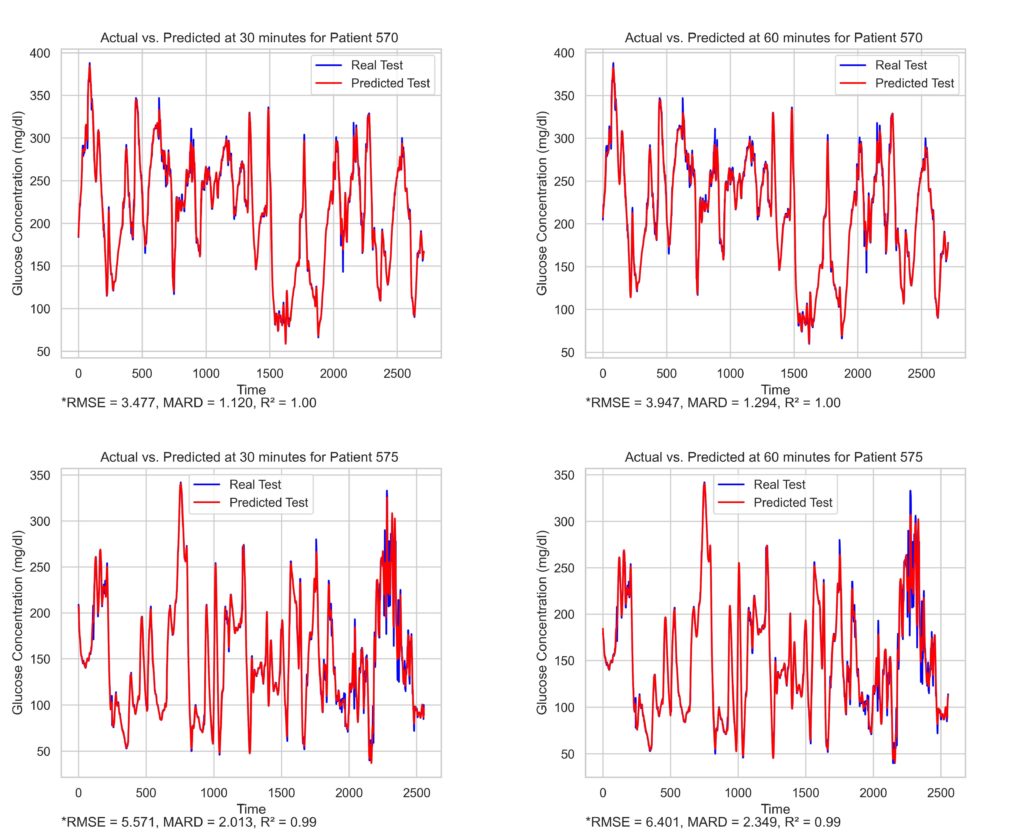
The findings suggest observable gains in predictive accuracy obtained using simple ML models, provided that the appropriate data-preparation mechanisms would be in place. This indicates that a simple and computationally lightweight model, such as MLR, could be used in, for instance, a mobile environment ‒ with positive results. The importance of handling missing data is also highlighted, as features having significant gaps hindered predictive accuracy. In such cases, a totally non-invasive feature configuration would most likely yield poor results. For these reasons, further research in this field is warranted, particularly within the context of using more complex models to identify any potential hidden links among non- invasive features.
References/Bibliography:
[1] W. Villena Gonzales, A. Mobashsher, and A. Abbosh, “The Progress of Glucose Monitoring—A Review of Invasive to Minimally and Non-Invasive Techniques, Devices and Sensors,” Sensors, vol. 19, no. 4, p. 800, Feb 2019.” Sensors, vol. 19, no. 4, p. 800, Feb 2019. [Online]. Available: http://dx.doi.org/10.3390/s19040800” http://dx.doi.org/10.3390/s19040800
[2] F. L. Schwartz, C. R. Marling, and R. C. Bunescu, “The Promise and Perils of Wearable Physiological Sensors for Diabetes Management,” Journal of Diabetes Science and Technology, vol. 12, no. 3, pp. 587–591, May 2018. [Online]. Available: https://doi. org/10.1177/1932296818763228” https://doi.org/10.1177/1932296818763228
[3] M. Gusev, L. Poposka, G. Spasevski, M. Kostoska, B. Koteska, M. Simjanoska, N. Ackovska, A. Stojmenski, J. Tasic, and J. Trontelj, “Noninvasive Glucose Measurement Using Machine Learning and Neural Network Methods and Correlation with Heart Rate Variability,” Hindawi Journal of Sensors, vol. 2020, pp. 1–13, January 2020. [Online]. Available: https://doi.org/10.1155/2020/9628281
[4] C. Marling and R. Bunescu, “The OhioT1DM Dataset for Blood Glucose Level Prediction: Update 2020,” Technical Report, 2020. [Online]. Available: http://smarthealth.cs.ohio.edu/bglp/OhioT1DM-dataset-paper.pdf
Student: Daniel Anthony Cilia
Course: B.Sc. IT (Hons.) Software Development
Supervisor: Dr. Michel Camilleri
Co-supervisor: Mr. Joseph Bonello
