Articles about modern advancements and discoveries in the medical industry are being published online on a daily basis. However, due to privacy issues, there is a lack of data available for building knowledge-extraction tools specialised in the medical industry. This research aims at creating a biomedical information-extraction tool called Remedi.
The system is specialised in identifying ‘medical problem’ and ‘treatment’ entities from unstructured text in the medical domain, as well as the relations between them. The tool consists of a biomedical named-entity recognition (BM-NER) model, as well as a biomedical relation-extraction (BM-RE) model. A subset of the i2b2 2010 challenge dataset [6] has been acquired to train the BM-NER and BM-RE components. The dataset includes annotated anonymised medical reports that could be used for concept extraction and relation- classification tasks.
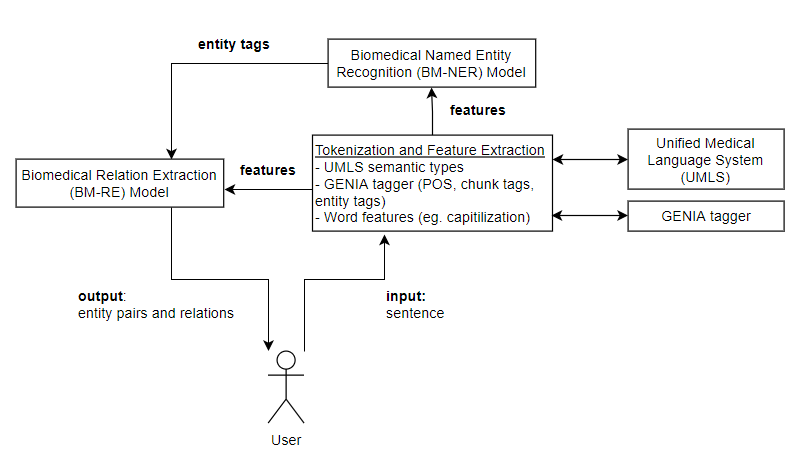
The research process revealed that most participants of the i2b2 concept-extraction challenge, which acquired the top results overall, employed conditional random field (CRF) models [6]. On the other hand, more recent research explores the use of neural networks, particularly, recurrent neural network (RNN) and convolutional neural network (CNN) models, which generated higher F-score results [7, 4]. Separate research explored the performance of a Bi-LSTM-CRF model on the dataset, which combined a bidirectional long-short-term-memory (Bi-LSTM) unit with a CRF layer for sequential tagging tasks, and obtained promising results [2]. However, Chalapathy et al. [2] did not mention the use of external sources (such as the Unified Medical Language System (UMLS) [1] and the GENIA tagger [5]) which has been noted to increase the performance of many models in the i2b2 challenge [6]. Hence, this research evaluates the impact of features extracted from external sources on the Bi-LSTM- CRF model, and compares it to the performance of a baseline CRF model using the same external features.
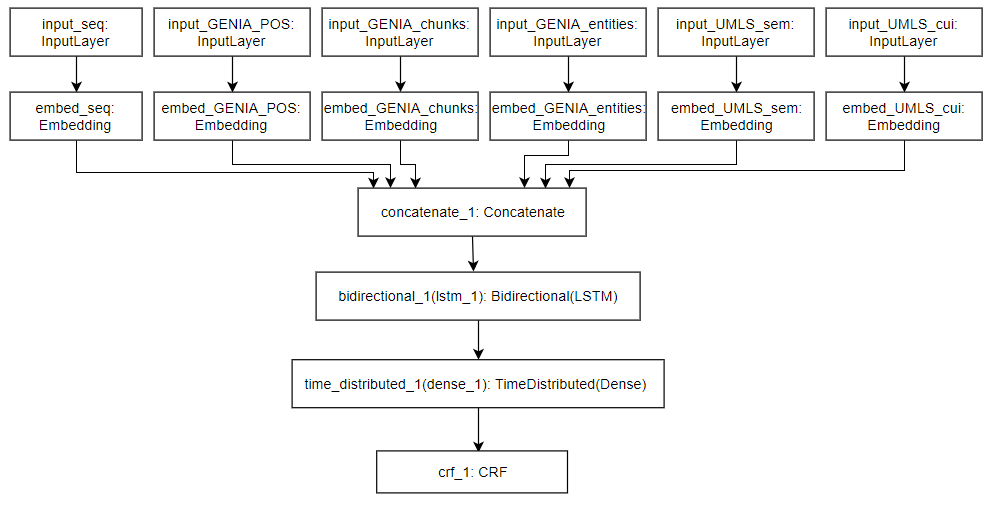
For the i2b2 relation-extraction challenge, the highest results were obtained using support vector machines (SVM) [6]. Research by Chikka et al [3] explored the use of Bi-LSTM models in comparison to constructed SVM models using a subset of the i2b2 dataset, which was also applied in this study. However, Chikka et al. [3] also do not make reference to the use of external sources. As a result, this study explores the performance of a Bi-LSTM model for relation- extraction using the same external sources employed in the experimentation for the BM-NER component, and comparing it to the models constructed in the research. This dissertation also carries out hyperparameter tuning for all implemented models, to determine which set of parameters would yield the best results for each model.
References/Bibliography:
[1] Olivier Bodenreider. The unified medical language system (umls): integrating biomedical terminology. Nucleic acids research, 32(suppl1): D267–D270, 2004.
[2] Raghavendra Chalapathy, Ehsan Zare Borzeshi, and Massimo Piccardi. Bidirectional lstm-crf for clinical concept extraction. arXiv preprint arXiv:1611.08373, 2016.
[3] Veera Raghavendra Chikka and Kamalakar Karlapalem. A hybrid deep learning approach for medical relation extraction. arXiv preprint arXiv:1806.11189, 2018.
[4] Zengjian Liu, Ming Yang, Xiaolong Wang, Qingcai Chen, Buzhou Tang, Zhe Wang,and Hua Xu. Entity recognition from clinical texts via recurrent neural network. BMC medical informatics and decision making, 17(2):67, 2017.
[5] Y Tsuruoka. Genia tagger: Part-of-speech tagging, shallow parsing, and named entity recognition for biomedical text. Available at: www-tsujii. is. su-tokyo. ac.jp/GENIA/tagger, 2006.
[6] Özlem Uzuner, Brett R South, Shuying Shen, and Scott L DuVall. 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association, 18(5):552–556, 2011.
[7] Yonghui Wu, Min Jiang, Jun Xu, Degui Zhi, and Hua Xu. Clinical named entity recognition using deep learning models. In AMIA Annual Symposium Proceedings, volume 2017, page 1812. American Medical Informatics Association, 2017.
Student: Valerija Holomjova
Course: B.Sc. IT (Hons.) Artificial Intelligence
Supervisor: Dr. Charlie Abela
